Genetic Modifications Leading To Antimicrobial Resistance
To understand why antibiotics must be used judiciously, the physician needs to understand how bacteria are able to adapt to their environment. Point mutations can develop in the Deoxyribonucleic acid of bacteria as they replicate.
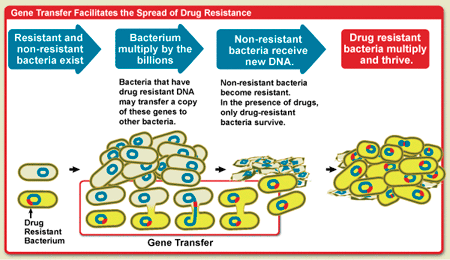
These mutations occur in the natural environment, but are of no survival advantage unless the bacteria are placed under selective pressures. In the case of a mutation that renders a bacterium resistant to a specific antibiotic, exposure to the specific antibiotic allows the bacterial clone that possesses the antibiotic resistance mutation to grow, while bacteria without the mutation die and no longer compete for nutrients. Thus the resistant strain becomes the dominant bacterial flora. In addition to point mutations bacteria can also use three major mechanisms to transfer genetic material among themselves:
- Conjugation. Bacteria often contain circular, double-stranded Deoxyribonucleic acid structures called plasmids. These circular Deoxyribonucleic acid structures lie outside the bacterial genome. Plasmids often carry resistance (“R”) genes. Through a mechanism called “conjugation,” plasmids can be transferred from one bacterium to another. The plasmid encodes for the formation of a pilus on the donor bacteria’s outer surface. The pilus attaches to a second bacterium and serves as bridge for the transfer of the plasmid Deoxyribonucleic acid from the donor to the recipient bacterium. Using this mechanism, a single resistant bacterium can transfer resistance to other bacteria.
- Transduction. Bacteriophages are protein-coated Deoxyribonucleic acid segments that attach to the bacterial wall and inject Deoxyribonucleic acid in a process called “transduction.” These infective particles can readily transfer resistance genes to multiple bacteria.
- Transformation. Donor bacteria can also release linear segments of chromosomal Deoxyribonucleic acid, which is then taken up by recipient bacteria and incorporated into the recipient’s genome. This process is called “transformation,” and the naked Deoxyribonucleic acid capable of incorporating into the genome of recipient bacteria is called a transposon. Natural transformation most commonly occurs in Streptococcus, Haemophilus, and Neisseria species. Transposons can transfer multiple antibiotic resistance genes in a single event and have been shown to be responsible for high-level vancomycin resistance in enterococci. Thus bacteria possess multiple ways to transfer their Deoxyribonucleic acid, and they promiscuously share genetic information. This promiscuity provides a survival advantage, allowing bacteria to quickly adapt to their environment.
About Antibiotic Resistance
- Bacteria can quickly alter their genetic makeup by a) point mutation. b) transfer of Deoxyribonucleic acid by plasmid conjugation. c) transfer of Deoxyribonucleic acid by bacteriophage transaction. d) transfer of naked Deoxyribonucleic acid by transposon transformation.
- The ability of bacteria to share Deoxyribonucleic acid provides a survival advantage, allowing them to quickly adapt to antibiotic exposure.
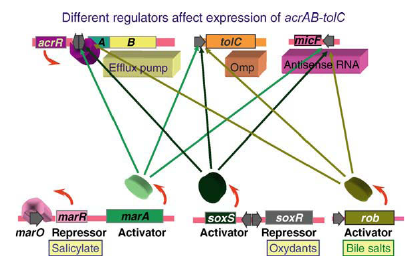
- Biochemical alterations leading to antibiotic resistance include a) degradation or modification of the antibiotic. b) reduction of the bacterial antibiotic concentration by inhibiting entry or by efflux pumps. c) modification of the antibiotic target.
- Under the selection pressure of antibiotics, the question is not whether, but when resistant bacteria will take over.
Biochemical Mechanisms For Antimicrobial Resistance
What are some of the proteins that these resistant genes encode for, and how do they work? The mechanisms by which bacteria resist antibiotics can be classified into three major groups:
- Degradation or modification of the antibiotic
- Reduction of the bacterial antibiotic concentration
- Modification of the antibiotic target
Degradation or Modification of the Antibiotic
β-Lactamases
Many bacteria synthesize one or more enzymes called β-lactamases that inactivate antibiotics by breaking the amide bond on the β-lactam ring. Transfer of β-lactamase activity occurs primarily through plasmids and transposons. Multiple classes of β-lactamases exist. Some preferentially break down penicillins; others preferentially destroy specific cephalosporins or carbenicillin. Extended-spectrum β-lactamases (ESBLs) readily destroy most cephalosporins. Another class of β-lactamase is resistant to clavulanate, an agent added to numerous antibiotics to inhibit β-lactamase activity. Some bacteria are able to produce β-lactamases called carbapenemases that are capable of inactivating imipenem and meropenem. Gram-negative bacilli produce a broader spectrum of β-lactamases than do gram-positive organisms, and therefore infections with gram-negative organisms more commonly arise in patients treated for prolonged periods with broad-spectrum antibiotics. In some instances, β-lactamase activity is low before the bacterium is exposed to antibiotics; however, following exposure, β-lactamase activity is induced. Enterobacter is a prime example. This gram-negative bacterium may appear sensitive to cephalosporins on initial testing. Following cephalosporin treatment, β-lactamase activity increases, resistance develops, and the patient’s infection relapses. For this reason, third-generation cephalosporins are not recommended for serious Enterobacter infections.
Other Enzyme Modifications of Antibiotics
Erythromycin is readily inactivated by an esterase that hydrolyzes the lactone ring of the antibiotic. This esterase has been identified in Escherichia coli. Other plasmid-mediated erythromycin inactivating enzymes have been discovered in Streptococcus species and S. aureus. Chloramphenicol is inactivated by chloram-phenicol acetyltransferase, which has been isolated from both gram-positive and gram-negative bacteria. Similarly, aminoglycosides can be inactivated by acetyltrans-ferases. Bacteria also inactivate this class of antibiotics by phosphorylation and adenylation. These resistance enzymes are found in many gram-negative strains and are increasingly detected in entero-cocci, S. aureus and S. epidermidis.
Reduction of the Bacterial Antibiotic Concentration
Interference with Antibiotic Entry
For an antibiotic to work, it must be able to penetrate the bacterium and reach its biochemical target. Gram-negative bacteria contain an outer lipid coat that impedes penetration by hydrophobic reagents (such as most antibiotics). The passage of hydrophobic antibiotics is facilitated by the presence of porins — small channels in the cell walls of gram-negative bacteria that allow the passage of charged molecules. Mutations leading to the loss of porins can reduce antibiotic penetration and lead to antibiotic resistance.
Production of Efflux Pumps
Transposons have been found that encode for an energy-dependent pump that can actively pump tetracycline out of bacteria. Active efflux of antibiotics has been observed in many enteric gram-negative bacteria, and this mechanism is used to resist tetracycline, macrolide, and fluoroquinolone antibiotic treatment. S. aureus, S. epidermidis, S. pyogenes, group В streptococci, and S. pneumoniae also can utilize energy-dependent efflux pumps to resist antibiotics.
Modification of the Antibiotic Target
Alterations of Cell Wall Precursors
Alteration of cell wall precursors is the basis for vancomycin-resistant Enterococcus. Vancomycin and teicoplanin binding requires that D-alanine-D-alanine be at the end of the peptidoglycan cell wall precursors of gram-positive bacteria. Resistant strains of Enterococcus faecium and Enterococcus faecalis contain the vanA plasmid, which encodes a protein that synthesizes D-alanine-D-lactate instead of D-alanine-D-alanine at the end of the peptidoglycan precursor. Loss of the terminal D-alanine markedly reduces vancomycin and teicoplanin binding, allowing the mutant bacterium to survive and grow in the presence of these antibiotics.
Changes in Target Enzymes
Penicillins and cephalosporins bind to specific proteins called penicillin-binding proteins in the bacterial cell wall. Penicillin-resistant S. pneumoniae demonstrate decreased numbers of penicillin-binding proteins or penicillin-binding proteins that bind penicillin with lower affinity, or both. Decreased penicillin binding reduces the ability of the antibiotic to kill the targeted bacteria.
The basis for antibiotic resistance in methicillin-resistant Staphylococcus aureus is production of a low affinity penicillin-binding protein encoded by the mecA gene. Mutations in the target enzymes dihydropteroate synthetase and dihydrofolate reductase cause sulfon-amide and trimethoprim resistance respectively. Single amino-acid mutations that alter Deoxyribonucleic acid gyrase function can result in resistance to fluoroquinolones.
Alterations in Ribosomal Binding Site
Tetracyclines, macrolides, lincosamides, and amino-glycosides all act by binding to and disrupting the function of bacterial ribosomes. A number of resistance genes encode for enzymes that demethylate adenine residues on bacterial ribosomal RNA, inhibiting antibiotic binding to the ribosome. Ribosomal resistance to gentamicin, tobramycin, and amikacin is less common because these aminoglycosides have several binding sites on the bacterial ribosome and require multiple bacterial mutations before their binding is blocked.
Conclusions
Bacteria can readily transfer antibiotic resistance genes. Bacteria have multiple mechanisms to destroy antibiotics, lower the antibiotic concentration, and interfere with antibiotic binding. Under the selective pressures of prolonged antibiotic treatment, the question is not whether, but when resistant bacteria will take over.